Incorporation of Artificial Intelligence and Epigenetic Genomics to verify Bladder Cancer DNA Methylation Biomarkers
 |
 |
|
Tun-Wen Pai, Professor and Department Chair Department of Computer Science and Information, National Taipei University of Technology Expertise : Bioinformatics, bio-medical informatics, medical imaging analysis |
Ming-Che Liu, Director Urology Department, Taipei Medical University Hospital Expertise : Urology Oncology . Clinical trial |
Contents
This project applies AI technologies to integrate Taiwan's Health Insurance Research Database (NHIRD), disease-gene association database, and DNA methylation profiling to develop an intelligent platform that can accelerate the discovery of novel and effective biomarkers for specific groups. Non-invasive collections of liquid biopsy of urine from patients with bladder cancer and/or urinary related diseases are the major specimens for DNA methylation experiments. Integration of primary and secondary biomarkers incorporating with deep learning technologies are designed for precision biomarker selection, and the designed toolkit is suitable for specific groups for early bladder cancer diagnosis and precision prognosis management.
Expected result
1. Developing a biomarker screening platform for selecting a set of effective DNA methylation biomarkers based on AI deep learning modules.
2. Performing pre-clinical verification experiments and establishing a risk assessment mechanism to confirm whether the designed DNA methylation biomarker testing toolkit meets clinical needs.
Benefits to industry
This project provides a non-invasive DNA methylation gene biomarker testing toolkit for bladder cancer patients under reduced risk and low cost requirements through urine liquid biopsy experiments. It is estimated that the number of bladder cancer patients in Taiwan will reach to 270,000 by 2035. The customized testing toolkits possess commercial advantages and are useful for bladder cancer patient groups at early detection or precision prognosis healthcare management.
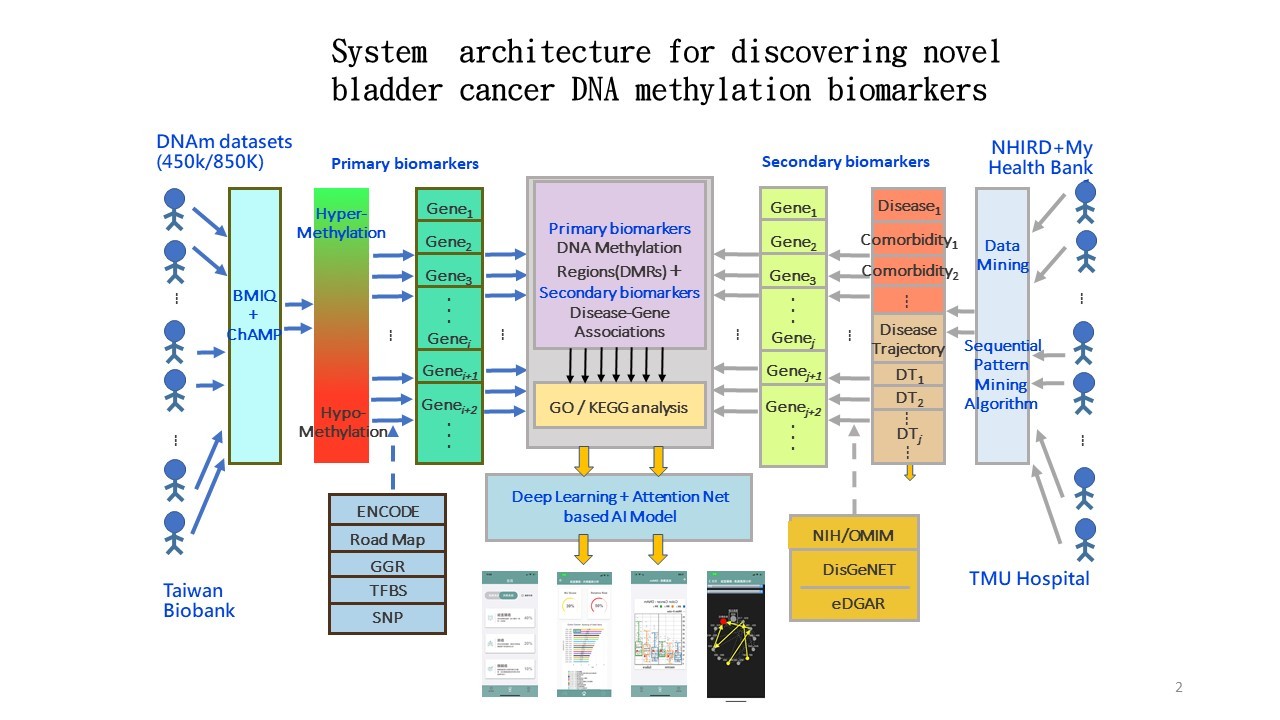
System architecture and platform for precision DNA methylation biomarker selection
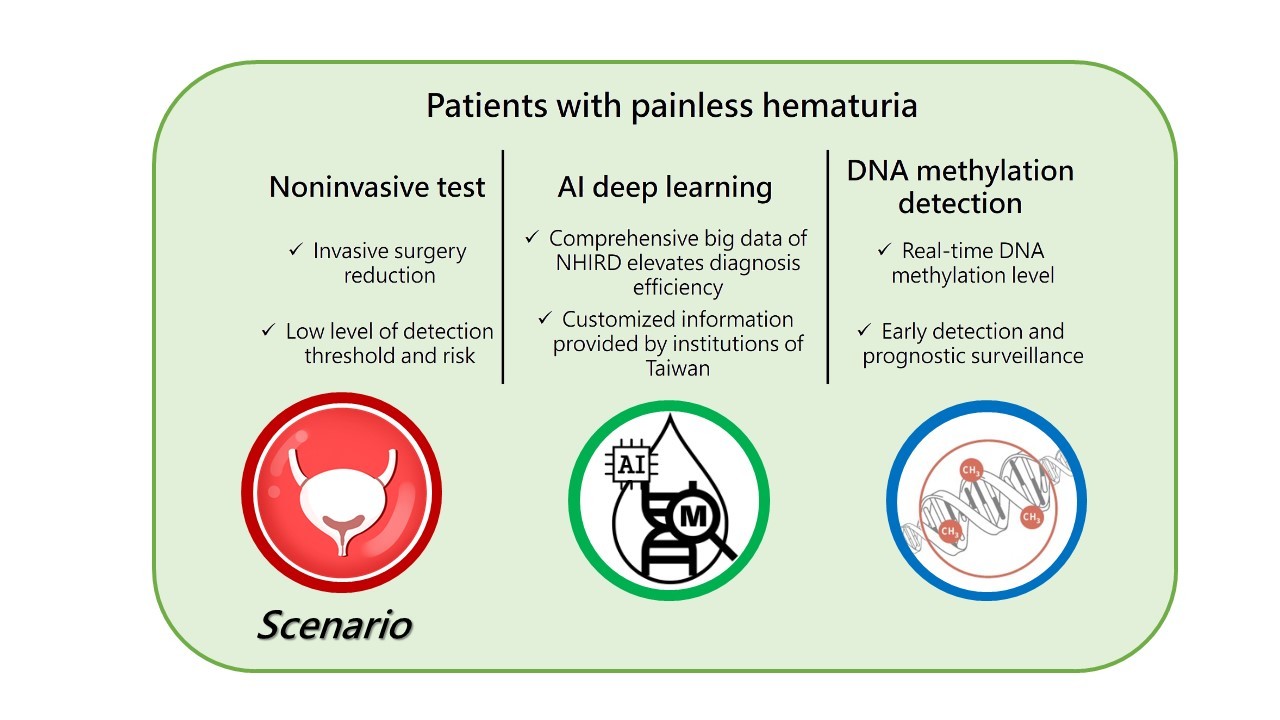
The high invasive cystoscope as the golden standard for preoperative diagnosis of patients with painless hematuria and prognostic surveillance of bladder cancer patients. Simultaneously, the invasive, painful, and uncomfortable genital examination of cystoscope brings inconvenience for patients. Therefore, we aim to identify the DNA methylation biomarkers for bladder cancer detection of Taiwanese via Taiwanese National Health Insurance Research Database (NHIRD) using AI deep learning appraoches. Further, through evaluating the DNA methylation profiles in urine specimens, the potential DNA methylation biomarkers in preoperative diagnosis and prognostic surveillance for bladder cancer will be identified.
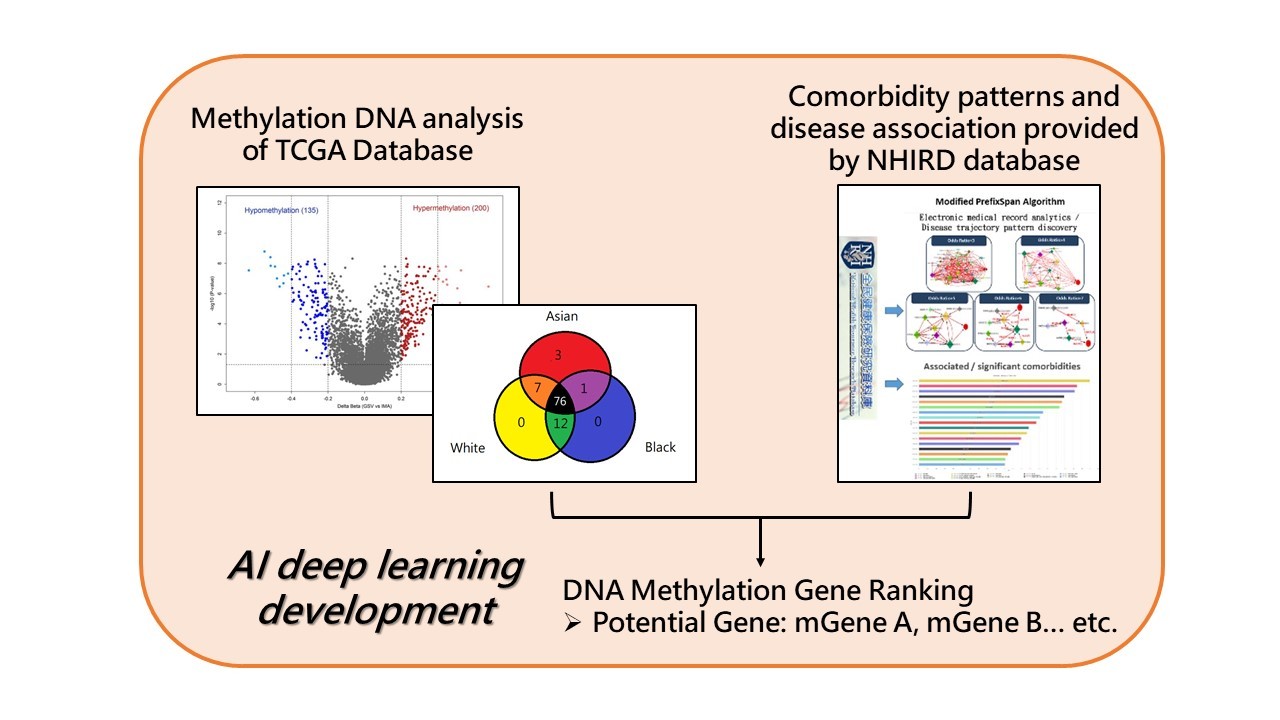
The research project applies AI deep learning model, integrated by DNA methylation data from The Cancer Genome Atlas (TCGA) database and disease-gene associations computed via Taiwan National Health Insurance Research Database (NHIRD), to identify the potential methylation biomarkers for specific groups of bladder cancer patients in Taiwan.
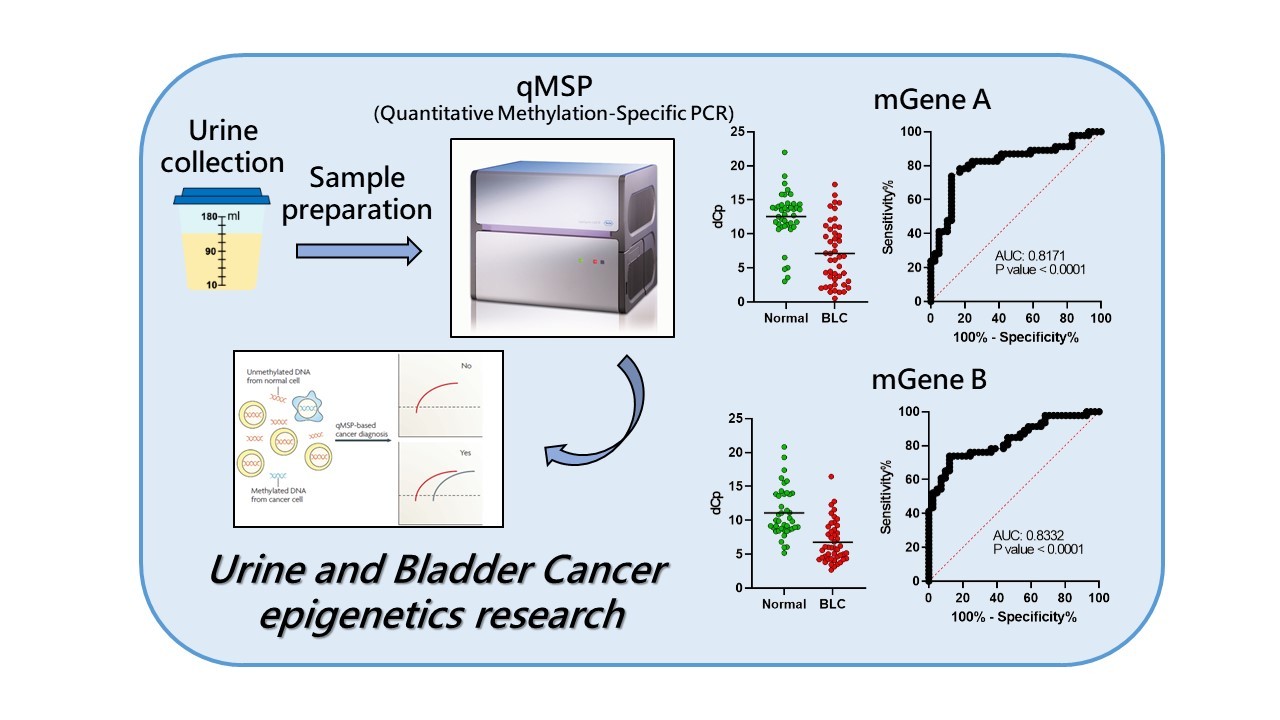
In the research project, urine (hematuria) specimens collected from bladder cancer and genitourinary disorder patients serve as the major source of liquid biological samples. Through integration of DNA methylation datasets from TCGA and comorbidity patterns and disease-gene associations provided by NHIRD database, the aberrant methylated DNA in the urine for bladder cancer will be identified. The training test, quantitative methylation-specific PCR (qMSP), will be used to identify appropriate biomarkers for bladder cancer early detection and prognostic surveillance for precision healthcare management.